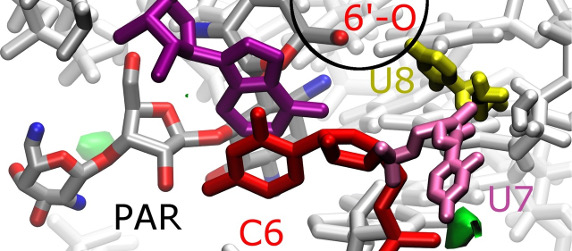
Predicting affinity of ligands to RNA. The case of an aminoglycoside-sensing riboswitch.
A synthetic riboswitch that binds aminoglycoside antibiotics was found to have regulatory activity while incorporated into cellular mRNA. This riboswitch is a flexible 27 nucleotide long RNA fragment whose atomistic three-dimensional structure in the complex with aminoglycosides was determined by nuclear magnetic resonance spectroscopy. However, the specificity of this riboswitch towards different aminoglycosides needs to be understood to be able to design a riboswitch variant possessing better regulatory properties against only one certain aminoglycoside.
The main aim of this project is to determine the dynamics of a synthetic aminoglycoside-sensing riboswitch to understand the aminoglycoside recognition mode and the structural basis of the riboswitch activity. We will also predict the structure of aminoglycoside-free riboswitch. The resulting goal is to determine the binding affinities of aminoglycosides to this riboswitch and its variants. This will allow us to propose aminoglycoside and riboswitch modifications to improve the regulation of gene expression.
The project is in collaboration with prof. Yuji Sugita and his group in RIKEN, Japan. Prof. Sugita is a Chief Scientist in RIKEN leading two laboratories: Theoretical Molecular Science Laboratory in Wako and Computational Biophysics Research Team in Kobe.
Principal Investigator: Prof. dr hab. Joanna Trylska, CeNT UW
Project period: 2018 – 2023
Funding: National Science Centre, HARMONIA
![]()
