Work package 1 (Centre of New Technologies, University of Warsaw)
“Positional and orientational steering of proteins toward biological membranes. Brownian dynamics approach”
MEMBDOC is a rigid-body Brownian dynamics software package designed for simulations of proteins’ diffusion near biological membranes.
Different modelling concepts can be used in MEMBDOC simulations of protein(s)-membrane systems. Implicit model describes the membrane as an impenetrable, planar, charged (with a continuous density) surface. With such a model one can observe electrostatic effects on proteins’ diffusion such as preferred orientations of proteins near the membrane and the influence of the membrane on protein-protein interactions in multi-particle systems. Modulation of the surface charge density corresponds to changes (in an average sense) in the composition of a lipid bilayer. More elaborate models, either coarse-grained or atomistic, allow one to study the effects of variations of discrete distributions of membrane charges or to introduce explicit representations of membrane-embedded proteins.
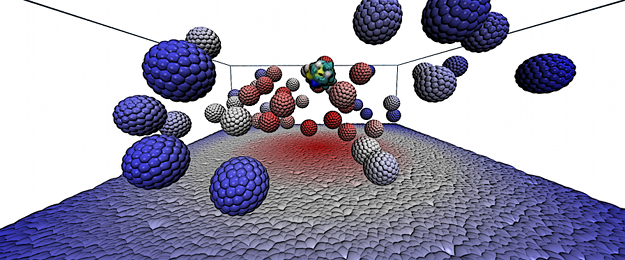
MEMBDOC is written in C and distributed as source code. Its main binary can be run either in a serial mode or in parallel, either on shared-memory machines using OpenMP and MPI libraries, or on architectures with distributed memory using the MPI library.
Work package 2 (University of Bergen)“De novo design of peptides as PR3 membrane binding inhibitors”
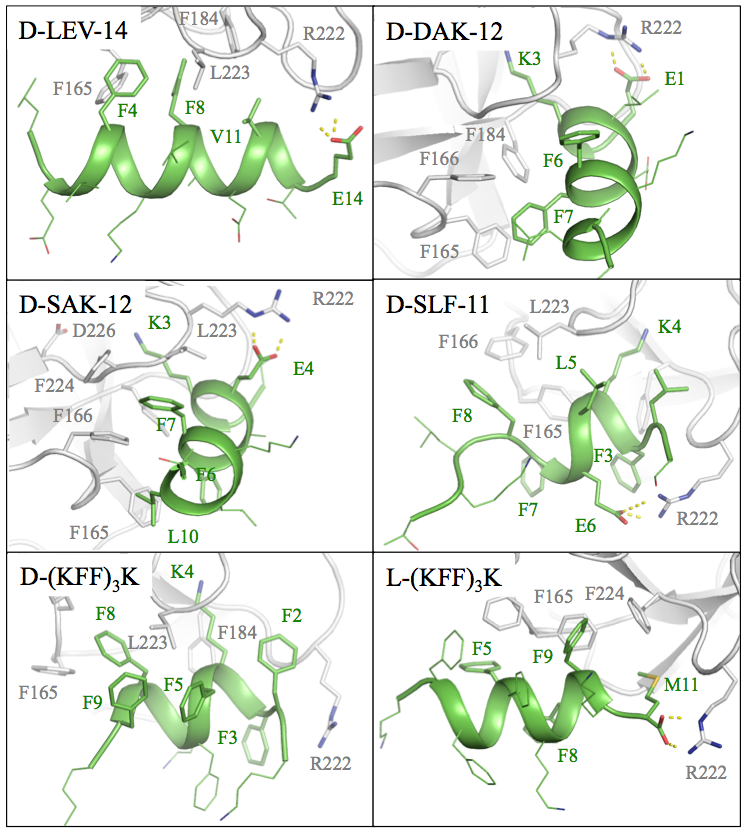
Applying docking and full-atom molecular dynamics simulations we designed nearly forty D-peptides targeting the PR3 membrane binding site. The sequences that we designed avoided the consensus sequence of typical PR3 ligands and did not target the active site of PR3 but exclusively the membrane interfacial binding site (IBS). We used at least two Phe residues in each sequence (to obtain π-π stacking interactions with corresponding Phe residues in the PR3 IBS) and engage the peptides in hydrophobic interactions with the PR3 IBS. Furthermore, upon optimizing the peptides, we combined large hydrophobic residues (such as Phe and Leu) with smaller ones (such as Val and Ala) to optimize the shape complementarity with the PR3 IBS surface. Another important consideration was to prevent the peptide binding to membranes. Since the cell membranes are negatively charged, we added acidic residues to the peptide sequences. Such peptides should also have better solubility.
Computational rational design of D-peptide sequences was accompanied with docking these peptides to PR3 IBS and followed by all-atom molecular dynamics simulations in explicit solvent. Finally, we selected six peptides that had promising poses and comparable strong predicted affinities. These were subjected to in vitro experiments in WP3.
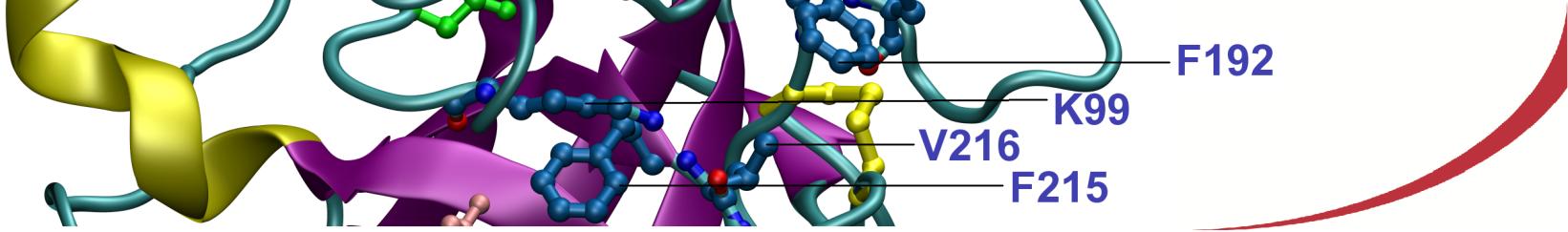
Tom Venken, Anne-Sophie Schillinger, Edvin Fuglebakk, Nathalie Reuter, Interactions stabilizing the C-terminal helix of human phospholipid scramblase 1 in lipid bilayers: A computational study, Biochimica et Biophysica Acta (BBA) – Biomembranes, 1859(7):1200-1210, 2017. https://doi.org/10.1016/j.bbamem.2017.03.019
Ksenia Maximova, Tom Venken, Nathalie Reuter, Joanna Trylska, D-Peptides as inhibitors of PR3-membrane interactions, Biochimica et Biophysica Acta (BBA) – Biomembranes, 1860:458-466, 2018. https://doi.org/10.1016/j.bbamem.2017.11.001
Ksenia Maximova, Nathalie Reuter, Joanna Trylska, Peptidomimetic inhibitors targeting the membrane-binding site of the neutrophil proteinase 3, BBA – Biomembranes, 1861:1502-1509, 2019. https://doi.org/10.1016/j.bbamem.2019.06.009
Work package 3 (Centre of New Technologies, University of Warsaw)
“In vitro measurements of the affinity of the compounds for PR3″
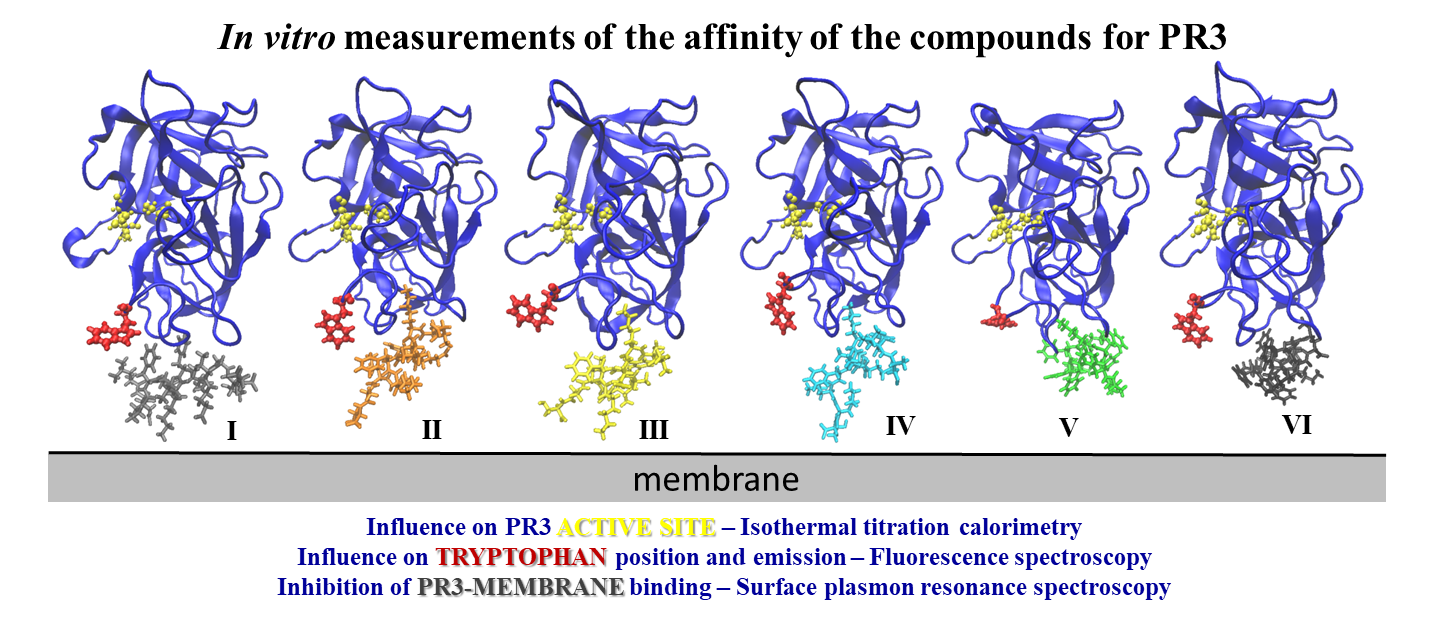
Six peptides, which had promising docking poses and comparable strong predicted binding affinities with PR3, were subjected to in vitro experiments.
We used surface plasmon resonance spectroscopy to verify if the binding of PR3 to lipids is inhibited by the designed D-peptides. The results showed no alteration of PR3-membrane binding by the peptides I, II and IV. The peptides III, V, and VI interfered with the PR3 binding to the membrane, however, the peptides V and VI have to be modified to prevent their interaction with the lipids.
We used isothermal titration calorimetry (ITC) to verify that the designed peptides do not affect the catalytic activity of PR3. Therefore, the peptide-inhibitors are different from the PR3 substrates and do not bind to the active site of PR3.
We used fluorescence spectroscopy to check the changes in the emission of PR3 tryptophans (TRP) to specify the binding.
Thus we verified the binding of a few peptides to PR3 hydrophobic pockets and the peptide inhibition of the PR3 binding to lipids.
Ksenia Maximova, Joanna Trylska, Kinetics of trypsin catalyzed hydrolysis determined by isothermal titration calorimetry, Analytical Biochemistry, 486:24-34, 2015. https://doi.org/10.1016/j.ab.2015.06.027
Ksenia Maximova, Tom Venken, Nathalie Reuter, Joanna Trylska, D-Peptides as inhibitors of PR3-membrane interactions, Biochimica et Biophysica Acta (BBA) – Biomembranes, 1860:458-466, 2018. https://doi.org/10.1016/j.bbamem.2017.11.001
Ksenia Maximova, Nathalie Reuter, Joanna Trylska, Peptidomimetic inhibitors targeting the membrane-binding site of the neutrophil proteinase 3, BBA – Biomembranes, 1861:1502-1509, 2019. https://doi.org/10.1016/j.bbamem.2019.06.009
Funding



