Ribosomes are ribonucleoprotein assemblies that conduct the process of translation. They consist of two unequal subunits, small and large, which in bacteria are denoted 30S and 50S, respectively. The 30S subunit contains over 20 proteins (S1, S2, etc.), one ~1500 nucleotide long 16S RNA chain, and it binds mRNA. The 50S subunit is composed of over 30 proteins (L1, L2, etc.), 23S RNA (~2900 nucleotides), and 5S rRNA (120 nucleotides). The subunits associate through a network of intermolecular interactions and form a 70S ribosome.
Amino acids are brought to the ribosome by transfer RNAs (tRNAs). The ribosome comprises three binding sites for tRNAs: A – aminoacyl, P – peptidyl and E – exit. The A-site accepts the incoming “charged” aminoacylated tRNA, the P-site holds the tRNA with the emerging peptide chain, and the E-site holds the deacylated tRNA before it leaves the ribosome. Peptide bond synthesis occurs between the aminoacyl and peptidyl ends of A- and P-site tRNAs. After the reaction, tRNAs must cooperatively translocate to their new positions (P and E) with the advance of mRNA by exactly one codon. During this translocation stage tRNAs must move a significant distance of about 20 Å. The E-site tRNA is then ready to leave the ribosome, a new tRNA may bind in the A-site, and the cycle repeats.
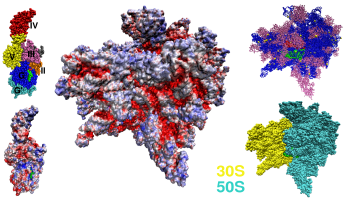
Figure: Left: Van der Waals sphere model of EF-G and its electrostatic potential calculated for the all-atom model (PDB entry 1FNM). Electrostatic potential is mapped on the molecular surface. The scale goes from red <= −5.0 kT/e, through white 0 kT/e to blue >= +5.0 kT/e. Middle: The electrostatic potential mapped on the molecular surface of the 70S ribosome [1,2]. Right: Upper inset shows the position of the A-site tRNA in green, P-site tRNA in red, rRNA in blue, and proteins in purple. Lower inset shows the orientation of the subunits.
Translation stages (initiation, elongation, and termination) are modulated by timely binding of various factors to the ribosome [3,4]. The elongation process involves two factors, EF-G and EF-Tu, which belong to the GTPase superfamily of proteins. EF-Tu-GTP complexes with the aminoacyl-tRNA (aa-tRNA) and delivers it to the A-site. When the appropriate charged tRNA is deposited into the A-site, the GTP is hydrolyzed, the aa-tRNA is accommodated, and the EF-Tu-GDP is released. Next, EF-G associates and promotes the translocation phase. This process is most likely facilitated by EF-G at the expense of GTP.
EF-Tu is a large (> 400-residue) protein with a net charge, near neutral pHs, of −16e for the T. thermophilus species. EF-Tu is composed of three domains G (or I), II and III. EF-Tu-GTP:aa-tRNA complex, called the ternary complex, is of elongated shape and is structurally related to EF-G. EF-G is a five domain protein with over 600 residues which exhibits a net negative charge of −22e. Both EF-G and ternary complex bind in the inter-subunit space of the ribosome and make contacts with the large and small subunits. There is also evidence that both EF-G and ternary complex undergo conformational change upon binding to the ribosome and upon GTP hydrolysis [3,4].
[1] M. M. Yusupov, G. Z. Yusupova, A. Baucom, K. Lieberman, T. N. Earnest, J. H. D. Cate, H. F. Noller, Science, 292:883, 2001.[2] K. Y. Sanbonmatsu, S. Joseph, and C-S. Tung, Proc. Natl. Acad. Sci. USA, 102:15854, 2005.
[3] A. Liljas. Structural Aspects of Protein Synthesis. World Scientific Publishing Co. Pte. Ltd., 2004.
[4] J. Nilsson and P. Nissen, Curr. Opin. Str. Biol., 15:349, 2005.
