RedMDStream
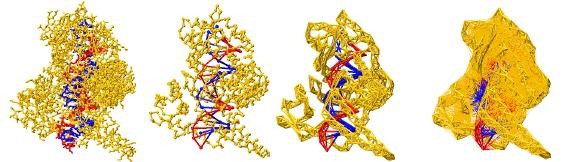
RedMD (Reduced Molecular Dynamics) is an open source package for molecular dynamics (MD) simulations and normal mode analysis of coarse-grained models of proteins and nucleic acids. Force fields are based on one-bead elastic network models and their extensions. RedMDStream extends the functionality of RedMD to allow efficient development and automatic parameterization of new coarse-grained models for proteins, nucleic acids and their complexes.
MINT
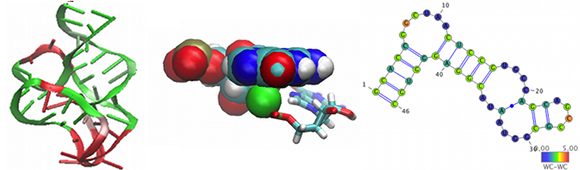
MINT (Motif Identifier for Nucleic Acids Trajectory) is an automatic tool for analyzing nucleic acids three-dimensional structures and molecular dynamics trajectories, including RNA hydrogen network pattern, stacking interactions and structural motifs such as helices, junctions and all kinds of loops. For the program, its documentation please visit MINT webserver page (http://mint.cent.uw.edu.pl).
RiboScanner

RiboScanner (http://riboscanner.cent.uw.edu.pl) is a server to analyze various physico-chemical properties of E. coli ribosomal RNA. Specifically, ribosomal RNA stretches can be verified as targets for sequence-specific inhibition by oligonucleotides. Further, peptide nucleic acid (PNA) oligomer sequences can be verified as complementary targets toward ribosomal RNA of E. coli as well as verified in terms of mismatches with human and mitochondrial rRNAs.
Surface Diver
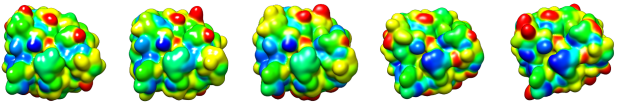
Surface Diver allows describing, comparing, and classifying proteins according to their physicochemical properties without the need of prior 3D structural alignment.
PDC
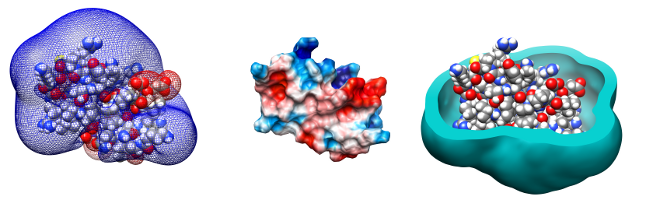
PDC (Potential Derived Charges) is a package to calculate effective charges for macromolecules so that they reproduce the molecular electrostatic potential calculated from the numerical solution of the Poisson-Boltzmann equation.
ION
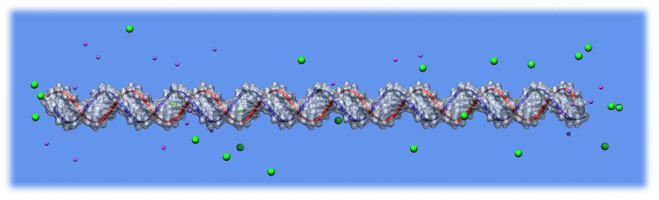
The ION software determines the distribution of ions around biomolecules using the Poisson model and Metropolis Monte-Carlo algorithm.
BD_BOX
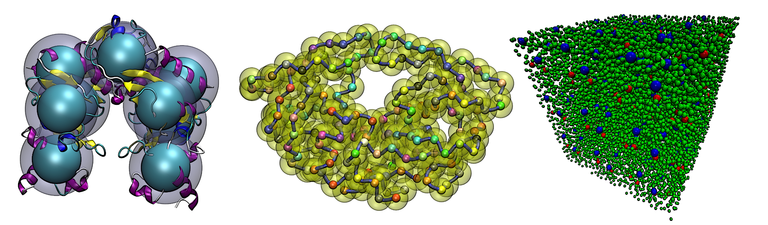
BD_BOX is an open source, scalable Brownian dynamics package for UNIX/LINUX platforms.
GeoStaS
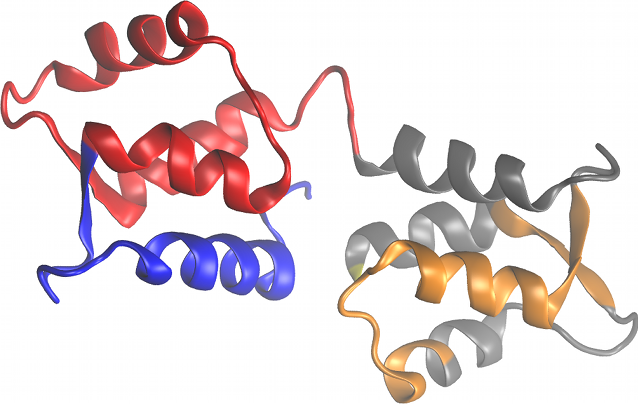
GeoStaS divides a biomolecule (proteins, nucleic acids and their complexes) into dynamic domains based on its different conformations, obtained from experiments or simulations.
